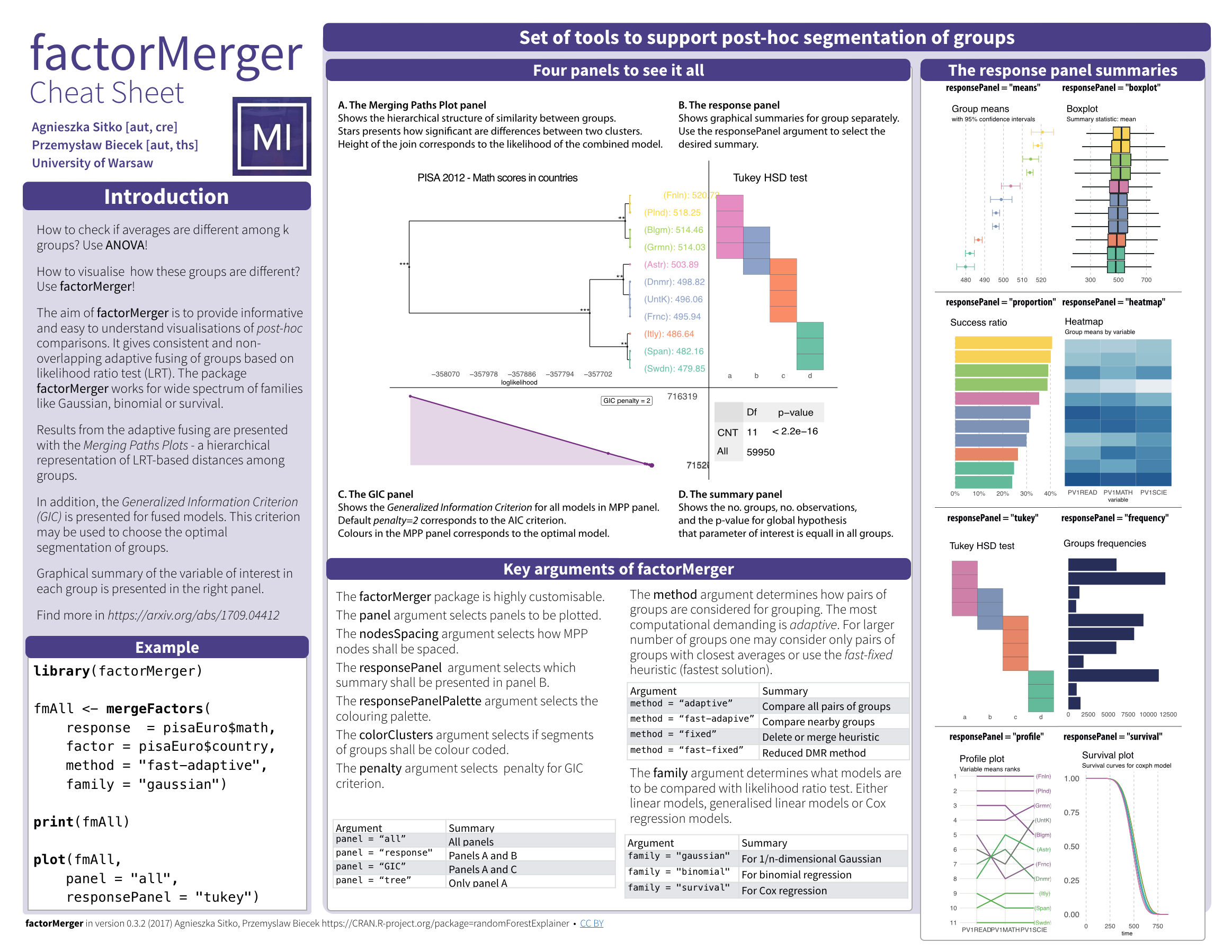
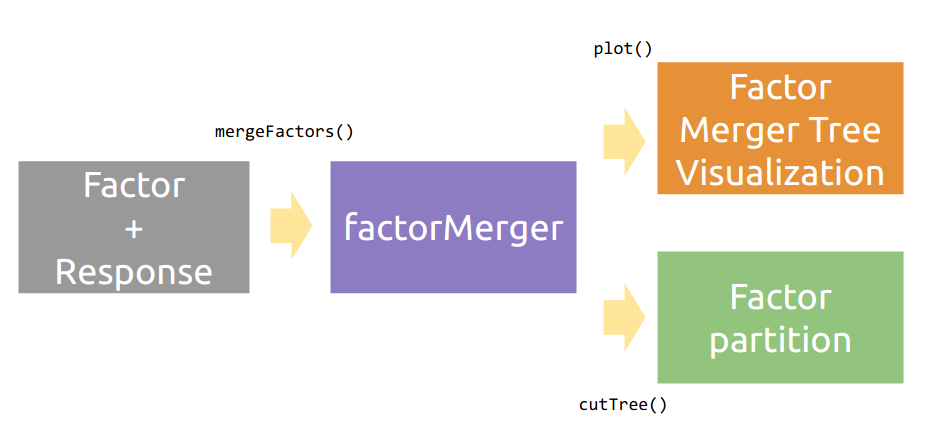
factorMerger: A Set of Tools to Support Adaptive Post-Hoc Fusing of Groups
The aim of this project is to create an algorithm of post-hoc testing that would enable to extract hierarchical structure of factors with respect to a given response.
Installing and loading factorMerger
factorMerger can be installed from CRAN as follows:
install.packages("factorMerger")
To install and load the latest version offactorMerger from Github run:
devtools::install_github("MI2DataLab/factorMerger", build_vignettes = FALSE)
Materials for factorMerger
A longer description of factorMerger may be found here (https://arxiv.org/abs/1709.04412).
Working with factorMerger
Examples
Survival analysis
library(factorMerger)
library(forcats) # distinguish meaningful factors (fct_lump)
data("BRCA")
brcaSurv <- survival::Surv(time = BRCA$time, event = BRCA$vitalStatus)
drugName <- fct_lump(as.factor(BRCA$drugName), prop = 0.05)
drugNameFM <- mergeFactors(response = brcaSurv[!is.na(drugName)],
factor = drugName[!is.na(drugName)],
family = "survival")
plot(drugNameFM, nodesSpacing = "effects", gicPanelColor = "grey2")