brainpy
A Python implementation of Baffling Recursive Algorithm for Isotopic distributioN calculations (BRAIN).
This is a direct translation of Han Hu's root-finding-free approach.
Documentation: http://mobiusklein.github.io/brainpy
Theoretical isotopic patterns appear when you can resolve distinct isotopes of an ion in a mass spectrum. Being able to predict the isotopic pattern of a molecule is useful for interpreting mass spectra to avoid counting the same ion with extra neutrons twice, recognizing the monoisotopic peak of a large multiply charged ion, or for discriminating among different elemental compositions of similar masses.
BRAIN takes an elemental composition represented by any Mapping-like Python object
and uses it to compute its aggregated isotopic distribution. All isotopic variants of the same
number of neutrons are collapsed into a single centroid peak, meaning it does not consider
isotopic fine structure.
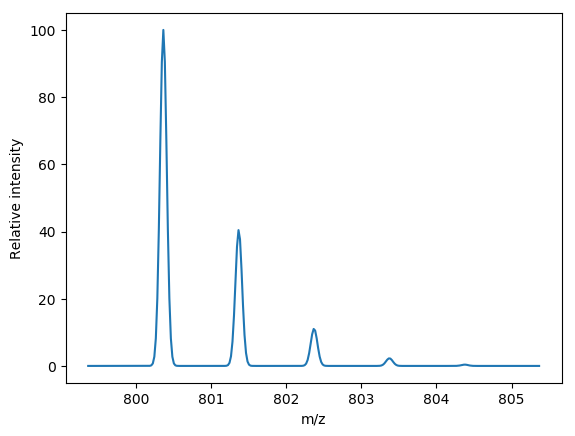
from brainpy import isotopic_variants
# Generate theoretical isotopic pattern
peptide = {'H': 53, 'C': 34, 'O': 15, 'N': 7}
theoretical_isotopic_cluster = isotopic_variants(peptide, npeaks=5, charge=1)
for peak in theoretical_isotopic_cluster:
print(peak.mz, peak.intensity)
# All following code is to illustrate what brainpy just did.
# produce a theoretical profile using a gaussian peak shape
import numpy as np
mz_grid = np.arange(theoretical_isotopic_cluster[0].mz - 1,
theoretical_isotopic_cluster[-1].mz + 1, 0.02)
intensity = np.zeros_like(mz_grid)
sigma = 0.002
for peak in theoretical_isotopic_cluster:
# Add gaussian peak shape centered around each theoretical peak
intensity += peak.intensity * np.exp(-(mz_grid - peak.mz) ** 2 / (2 * sigma)
) / (np.sqrt(2 * np.pi) * sigma)
# Normalize profile to 0-100
intensity = (intensity / intensity.max()) * 100
# draw the profile
from matplotlib import pyplot as plt
plt.plot(mz_grid, intensity)
plt.xlabel("m/z")
plt.ylabel("Relative intensity")Installing
brainpy has three implementations, a pure Python implementation, a Cython translation
of that implementation, and a pure C implementation that releases the GIL.
To install from a package index, you will need to have a C compiler appropriate to your Python version to build these extension modules. Additionally, there are prebuilt wheels for Windows available on PyPI.
$ pip install brain-isotopic-distribution
To build from source, in addition to a C compiler you will also need to install a recent version of Cython to transpile C code.
Original Algorithm:
P. Dittwald, J. Claesen, T. Burzykowski, D. Valkenborg, and A. Gambin, “BRAIN: a universal tool for high-throughput calculations of the isotopic distribution for mass spectrometry.,” Anal. Chem., vol. 85, no. 4, pp. 1991–4, Feb. 2013.
Original Implementation:
H. Hu, P. Dittwald, J. Zaia, and D. Valkenborg, “Comment on ‘Computation of isotopic peak center-mass distribution by fourier transform’.,” Anal. Chem., vol. 85, no. 24, pp. 12189–92, Dec. 2013.