bayerstraits_16s
This is a temp version of bayerstraits_16s
Introduction
- bayerstraits_16s infers traits by 16S
- input: otu table (-t) and otu sequences (-s)
- requirement: mafft
- Optional: fasttree
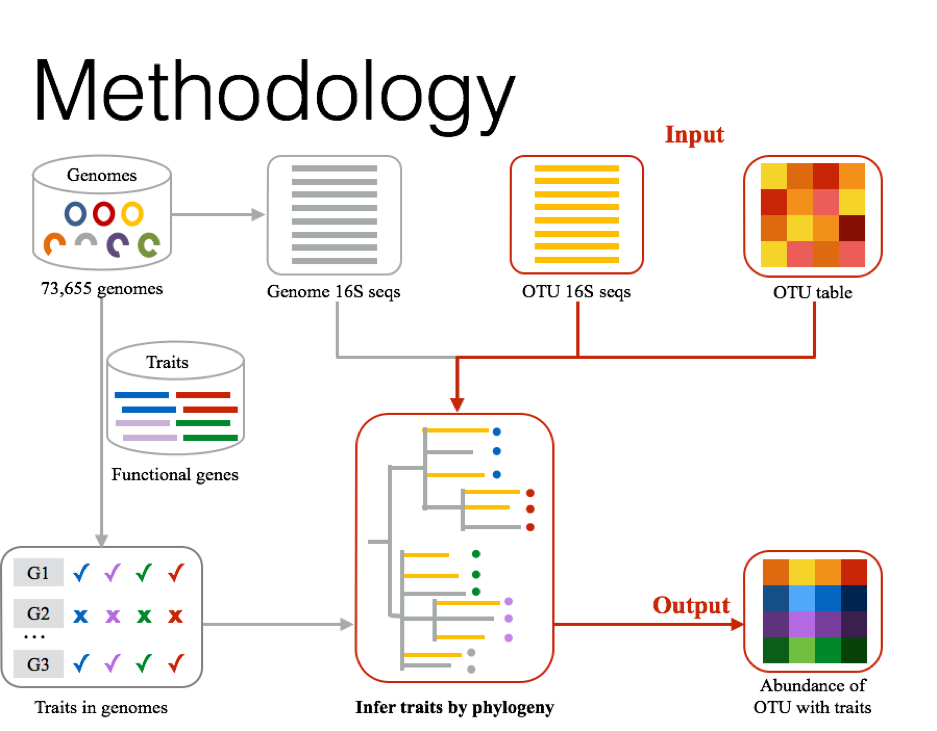
Install
pip install bayerstraits_16s
in preparation: anaconda download caozhichongchong/bayerstraits_16s
Test (any of these two commands)
bayerstraits_16s --test
bayerstraits_16s -t your.otu.table -s your.otu.seqs
Availability
in preparation: https://anaconda.org/caozhichongchong/bayerstraits_16s
https://pypi.org/project/bayerstraits_16s
How to use it
-
test the bayerstraits_16s
bayerstraits_16s --test -
try your data
bayerstraits_16s -t your.otu.table -s your.otu.seqs
bayerstraits_16s -t your.otu.table -s your.otu.seqs -top 2000 -
use your own traits
bayerstraits_16s -t your.otu.table -s your.otu.seqs --rs your.own.reference.16s --rt your.own.reference.traits
-
your.own.reference.16s is a fasta file containing the 16S sequences of your genomes
>Genome_ID1
ATGC...
>Genome_ID2
ATGC... -
your.own.reference.traits is a metadata of whether there's trait in your genomes (0 for no and 1 for yes)
Genome_ID1 0
Genome_ID1 1
Results
The result dir of "Bayers_model":
-
filename.infertraits.txt: the OTUs inferring as butyrate-producing bacteria (1.0, 0.5) and non-butyrate-producing bacteria (0.0). -
filename.infertraits.abu: the total abundance of butyrate-producing bacteria in all samples. -
filename.infertraits.otu_table: the otu_table of butyrate-producing bacteria in all samples.
The result dir of "Filtered_OTU":
- Some temp files of filtered OTUs, alignment, and tree.
Copyright
Copyright: An Ni Zhang, Prof. Eric Alm, Alm Lab in MIT
Citation: Not yet, coming soon!
Contact: anniz44@mit.edu