Job Dispatcher viewers is a TypeScript module that uses Fabric.js (based on a HTML canvas) to implement interactive visualisations, Visual Output and Functional Predictions. The app is provided as a ES module, can be used as plugin and also as Web Components. A Node.js CLI application is also provided to generate static figures in SVG and PNG formats.
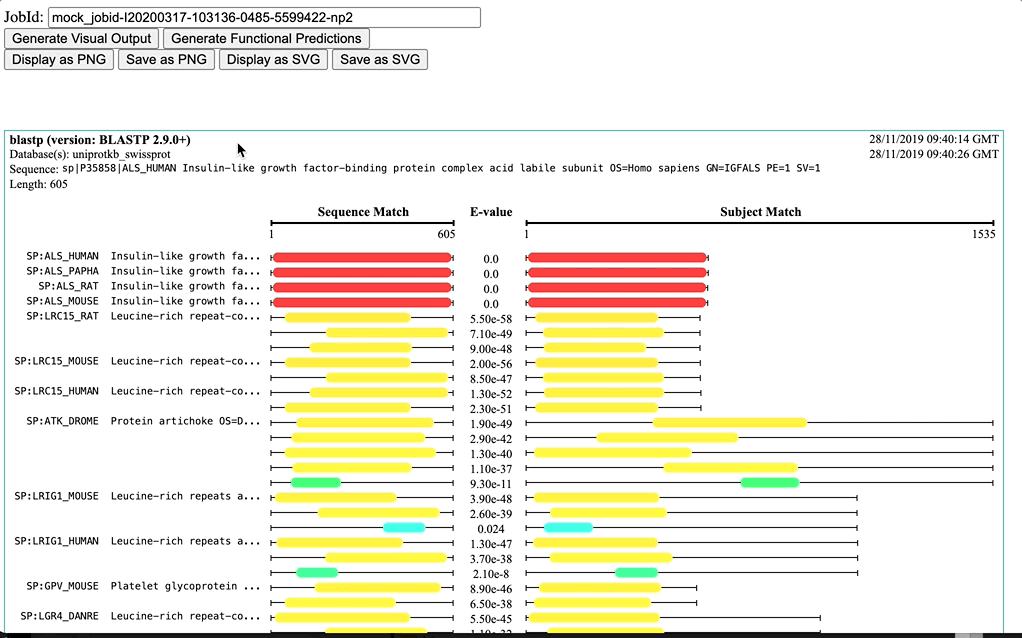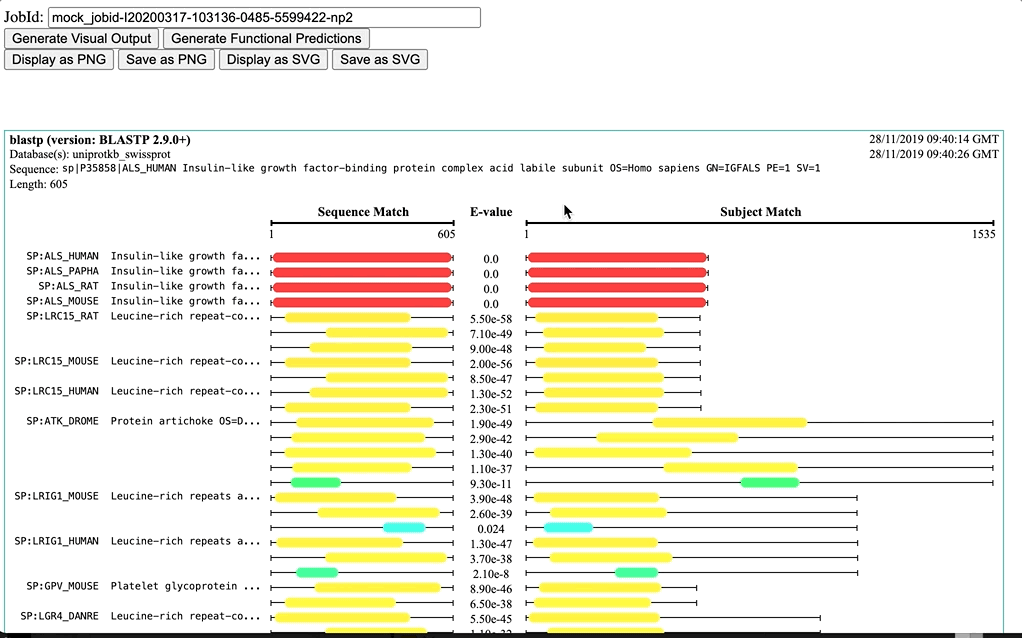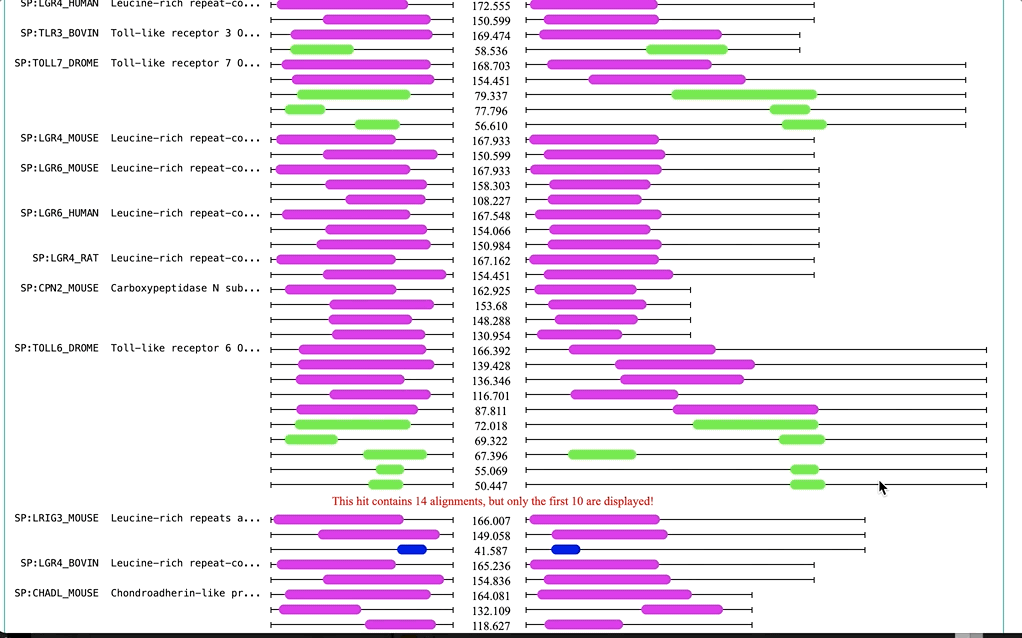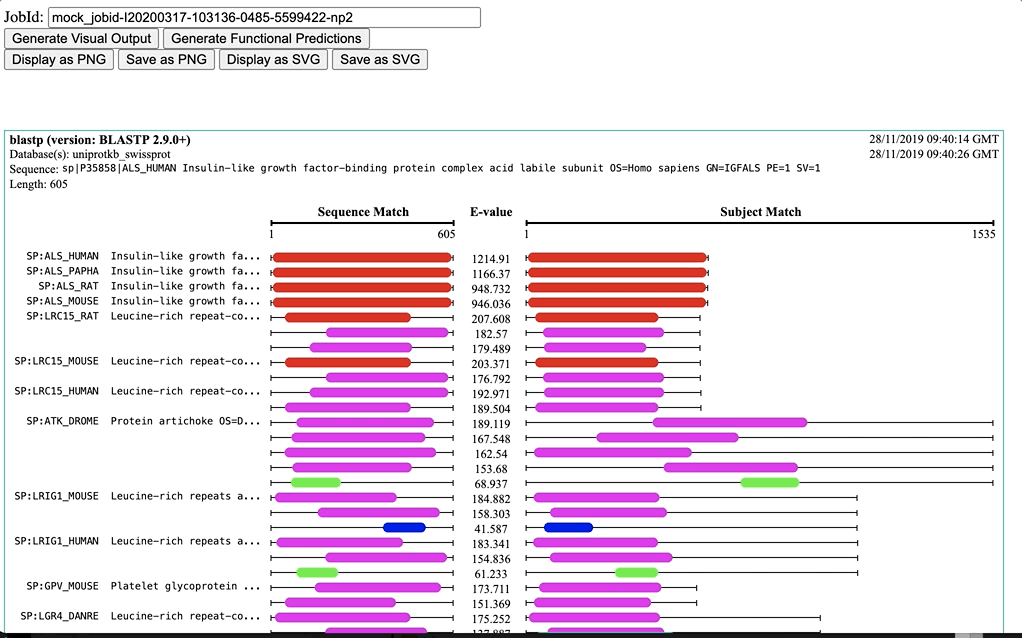
Visual Output is a diagram visualisation for the hit sequences of sequence similarity tools (e.g. NCBI BLAST+ and FASTA, that highlights the region of the sequences that is matching the query sequence and provides corresponding E-values. This currently works only with Job Dispatcher JobIDs, as it depends on the JSON output provided by the service.
Functional Predictions is a diagram of the domain ranges that compose hit sequences of sequence similarity tools (e.g. NCBI BLAST+ and FASTA. This visualisation gives a good overview of domain location vs hit/query sequence matching. Predictions are obtained from InterPro and cover several domain databases/resources, including: Pfam, SUPERFAMILY, SMART, CATH-Gene3D, CDD, etc.
Three colouring schemes are provided.
-
Dynamic: It is based on the range of E-values (from min to max)) -
Fixed: It a fixed scale for E-values (from 0.0 to 100.0) -
NCBI BLAST: It is a fixed scale based on NCBI-BLAST+ bit-score ranges (bit-score <40 to >=200)
Building the applications requires Node.js (tested with v16.16.0 and v18.7.0). Additional requirements, are downloaded and installed automatically with npm CLI. See full list of dependencies in package.json
During development, you can simply clone this repository and install all the dependencies required to run the webpack and webpack-dev-server:
npm installTo play with the development server (watches for file changes), in two terminal windows in parallel, run:
# runs a server at localhost:8080
npm run dev# builds the package and copies index.html over to ./dist
npm run dev2To compile the application for production, run:
npm run buildTo compile the CLI during development (watches for file changes), run:
npm run dev:cliTo compile the CLI for production, run:
npm run build:cliTest the CLI by simply running (or passing the options -h or --help):
node --es-module-specifier-resolution=node bin/jd-viewers-cli.jsTesting the CLI using a mock jobId, which loads data from files under ./src/testdata.
node --es-module-specifier-resolution=node bin/jd-viewers-cli.js vo -i mock_jobid-I20200317-103136-0485-5599422-np2 -o test.png -of png -vPassing the same local files:
node --es-module-specifier-resolution=node bin/jd-viewers-cli.js fp -i ./src/testdata/ncbiblast.json -ix ./src/testdata/iprmc.xml -o test.svg -of svg -vPassing a valid JobId (replace with a current JobId, as this one might have expired), where the data will be fetched and stored locally:
node --es-module-specifier-resolution=node bin/jd-viewers-cli.js fp -i ncbiblast-R20200602-114955-0302-11398737-np2 -o test.svg -of svg -vSome rendering options can be optionally passed to the CLI, including: -hits, number of hits;
-hsps, number of HSPs; and -color, color scheme. For example:
node --es-module-specifier-resolution=nodebin/jd-viewers-cli.js fp -i ncbiblast-R20200602-114955-0302-11398737-np2 -o test.svg -of svg -color 'ncbiblast' -hits 50 -vThis package has been published to NPM at @ebi-jdispatcher/jdispatcher-viewers.
# npm package under ebi-jdispatcher org
npm init --scope=@ebi-jdispatcher
# testing building the package
npm install
# publish to npm
npm publish --access publicIf you find any bugs or issues please log them in the issue tracker.
See CHANGELOG.md.
The European Bioinformatics Institute - EMBL-EBI, is an Intergovernmental Organization which, as part of the European Molecular Biology Laboratory family, focuses on research and services in bioinformatics.
Apache License 2.0. See LICENSE for details.