georeader is a package to process raster data from different satellite missions. georeader makes easy to read specific areas of your image, to reproject images from different satellites to a common grid (georeader.read), to go from vector to raster formats (georeader.vectorize and georeader.rasterize) or to do radiance to reflectance conversions (georeader.reflectance).
georeader is mainly used to process satellite data for scientific usage, to create ML-ready datasets and to implement end-to-end operational inference pipelines (e.g. the Kherson Dam Break floodmap).
The core package dependencies are numpy, rasterio, shapely and geopandas.
pip install georeader-spacemlRead from a Sentinel-2 image a fixed size subimage on an specific
lon,latlocation (directly from the S2 public Google Cloud bucket):
# This snippet requires:
# pip install fsspec gcsfs google-cloud-storage
import os
os.environ["GS_NO_SIGN_REQUEST"] = "YES"
from georeader.readers import S2_SAFE_reader
from georeader import read
cords_read = (-104.394, 32.026) # long, lat
crs_cords = "EPSG:4326"
s2_safe_path = S2_SAFE_reader.s2_public_bucket_path("S2B_MSIL1C_20191008T173219_N0208_R055_T13SER_20191008T204555.SAFE")
s2obj = S2_SAFE_reader.s2loader(s2_safe_path,
out_res=10, bands=["B04","B03","B02"])
# copy to local avoids http errors specially when not using a Google Cloud project.
# This will only copy the bands set up above B04, B03 and B02
s2obj = s2obj.cache_product_to_local_dir(".")
# See also read.read_from_bounds, read.read_from_polygon for different ways of croping an image
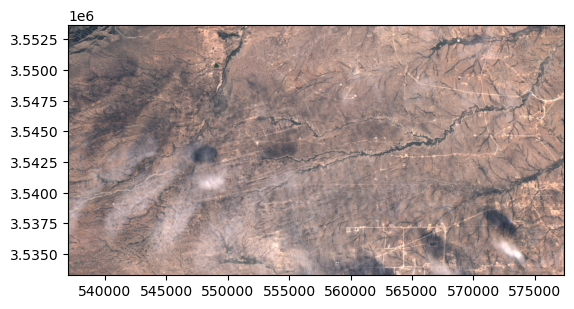
data = read.read_from_center_coords(s2obj,cords_read, shape=(2040, 4040),
crs_center_coords=crs_cords)
data_memory = data.load() # this loads the data to memory
data_memory # GeoTensor object>> Transform: | 10.00, 0.00, 537020.00|
| 0.00,-10.00, 3553680.00|
| 0.00, 0.00, 1.00|
Shape: (3, 2040, 4040)
Resolution: (10.0, 10.0)
Bounds: (537020.0, 3533280.0, 577420.0, 3553680.0)
CRS: EPSG:32613
fill_value_default: 0
In the .values attribute we have the plain numpy array that we can plot with show:
from rasterio.plot import show
show(data_memory.values/3500, transform=data_memory.transform)Saving the GeoTensor as a COG GeoTIFF:
from georeader.save import save_cog
# Supports writing in bucket location (e.g. gs://bucket-name/s2_crop.tif)
save_cog(data_memory, "s2_crop.tif", descriptions=s2obj.bands)-
Reading Sentinel-2 images from the public Google bucket
-
Explore metadata of Sentinel-2 object
-
Query Sentinel-2 images over a location and time span, mosaic and plot them
-
Sentinel-2 images from GEE and CloudSEN12 cloud detection
-
Tutorial to read overlapping tiles from a GeoTIFF and a Sentinel-2 image
- Example of reading a Proba-V image overlapping with Sentinel-2 forcing same resolution
-
Work with EMIT images
- Read overlapping images of PRISMA and EMIT
- Read EnMAP images, integrate them to Sentinel-2 bands, convert radiance to TOA reflectance and run CloudSEN12 cloud detection model
- georeader with ml4floods to automatically download and produce flood extent maps: the Kherson Dam Break example
- georeader with STARCOP to simulate Sentinel-2 from AVIRIS images
- georeader with STARCOP to run plume detection in EMIT images
- georeader with CloudSEN12 to run cloud detection in Sentinel-2 images
If you find this code useful please cite:
@article{portales-julia_global_2023,
title = {Global flood extent segmentation in optical satellite images},
volume = {13},
issn = {2045-2322},
doi = {10.1038/s41598-023-47595-7},
number = {1},
urldate = {2023-11-30},
journal = {Scientific Reports},
author = {Portalés-Julià, Enrique and Mateo-García, Gonzalo and Purcell, Cormac and Gómez-Chova, Luis},
month = nov,
year = {2023},
pages = {20316},
}
@article{ruzicka_starcop_2023,
title = {Semantic segmentation of methane plumes with hyperspectral machine learning models},
volume = {13},
issn = {2045-2322},
url = {https://www.nature.com/articles/s41598-023-44918-6},
doi = {10.1038/s41598-023-44918-6},
number = {1},
journal = {Scientific Reports},
author = {Růžička, Vít and Mateo-Garcia, Gonzalo and Gómez-Chova, Luis and Vaughan, Anna, and Guanter, Luis and Markham, Andrew},
month = nov,
year = {2023},
pages = {19999},
}
This research has been supported by the DEEPCLOUD project (PID2019-109026RB-I00) funded by the Spanish Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033) and the European Union (NextGenerationEU).