The Metabolic Disassembler
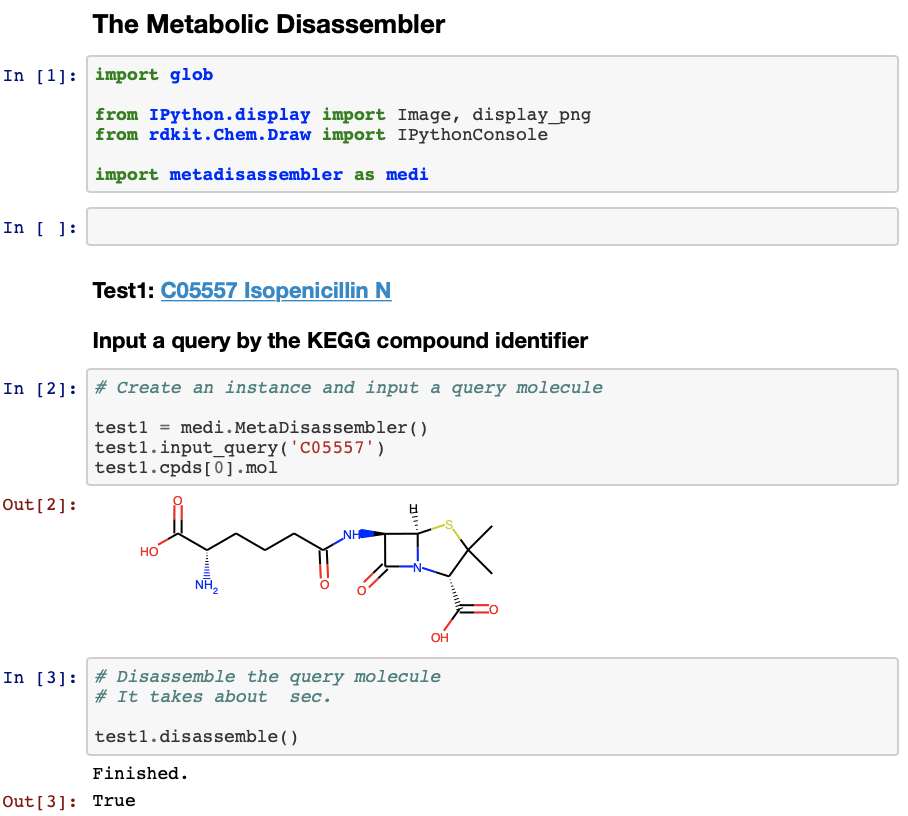
The Metabolic Disassembler is a Python package to automatically predict a combination of biosynthetic units in a natural product. This software would help to reveal the starting materials of the target natural product.
Installation
Install The Metabolic Disassembler with pip.
$ pip install metadisassemblerRequirements
- Python (3.6)
- RDKit (version 2019.09.2.0)
- NetworkX (version 2.2)
- CairoSVG (version 2.4.2)
- Pillow (PIL) (version 6.2.1)
- Pandas (version 0.25.3)
- Matplotlib (version 3.1.2)
Command Line Usage
% metadisassembler -h
usage: metadisassembler [-h] [-t TIME] [--hide] [-c] query
positional arguments:
query MDL_Molfile, SMILES, InChI, KEGG_COMPOUND_ID,
KNApSAcK_COMPOUND_ID
optional arguments:
-h, --help show this help message and exit
-t TIME, --time TIME set a time limit [s] [default: 300]
--hide hide stereochemistry [default: False]
-c, --color output color allocation information [default: False]Basic Usage
A usage example by Jupyter Notebook can be seen here.
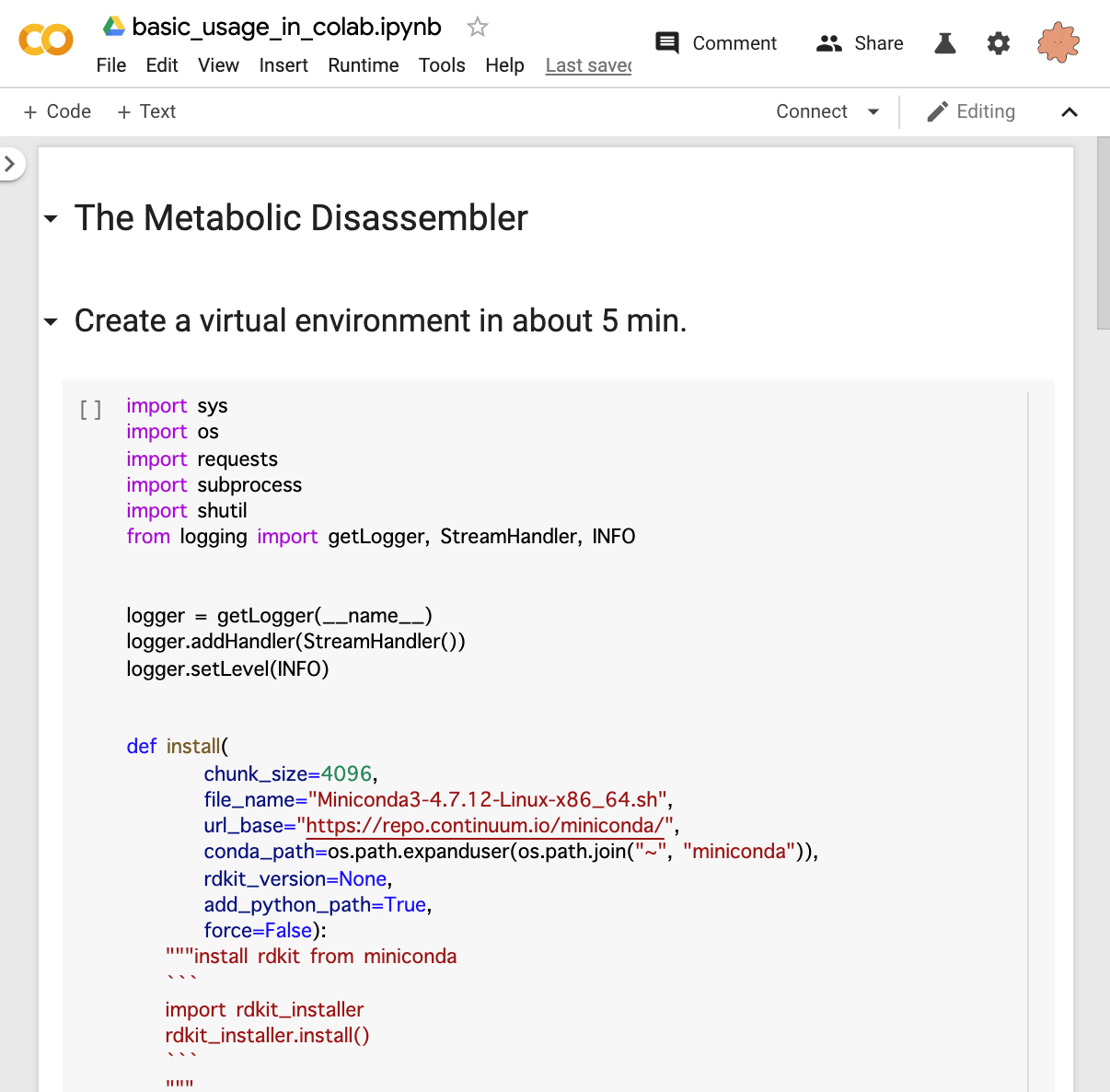
Google Colab
You can try the process from installation to the basic usage in Google Colab.
Citation
Amano K, Matsumoto T, Tanaka K, Funatsu K, Kotera M. Metabolic disassembler for understanding and predicting the biosynthetic units of natural products. BMC Bioinformatics 20, 728 (2019) doi:10.1186/s12859-019-3183-9