Python library for mutational spectra analysis
- python 3.8+
pip3 install pymutspechttps://pypi.org/project/PyMutSpec/
from Bio import SeqIO
from pymutspec.annotation import calculate_mutspec, CodonAnnotation
from pymutspec.draw import plot_mutspec12, plot_mutspec192
coda = CodonAnnotation(gencode=2) # mitochondrial genetic code
path_to_observed_mutations = ...
path_to_reference_seq = ...
# load data (mutations and sequence)
gene = SeqIO.parse(path_to_reference_seq, format='fasta')
observed_mutations = pd.read_csv(path_to_observed_mutations, sep='\t')
for col in ['Mut', 'MutType']:
assert col in observed_mutations.columns
# sample only syn mutations
mut_syn = observed_mutations[observed_mutations.MutType >= 1] # 0 for all mutations, 1 for syn, 2 for fourfold syn (syn4f)
# derive expected mutations from reference gene
sbs12_freqs, sbs192_freqs = coda.collect_exp_mut_freqs(gene, labels['all', 'syn', 'syn4f'])
sbs12_freqs_syn = sbs12_freqs['syn']
sbs192_freqs_syn = sbs192_freqs['syn']
# calculate mutation spectra
spectra12 = calculate_mutspec(mut_syn, sbs12_freqs_syn, use_context=False)
spectra192 = calculate_mutspec(mut_syn, sbs192_freqs_syn, use_context=True)
# plot mutation spectra
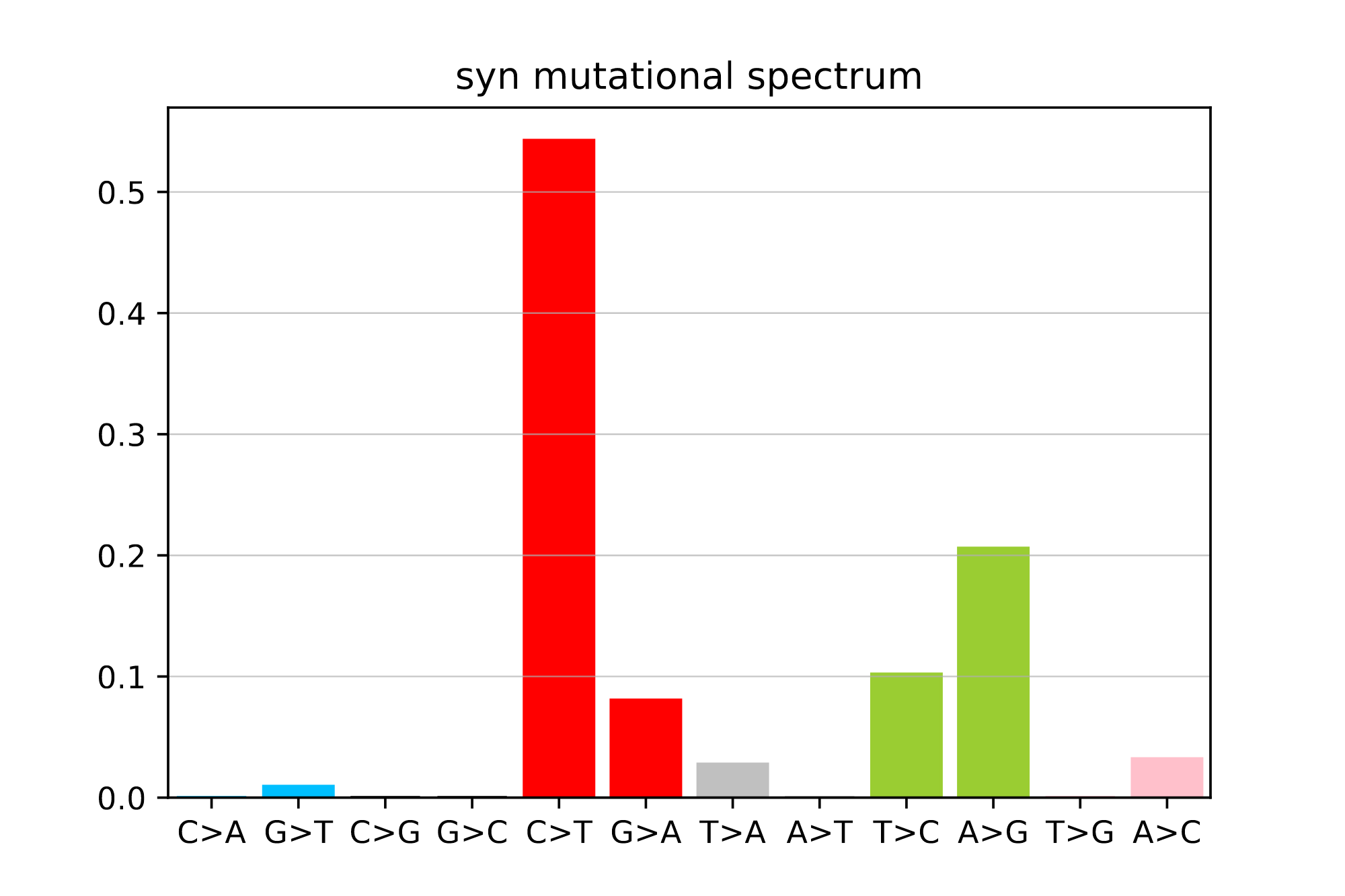
plot_mutspec12(spectra12)
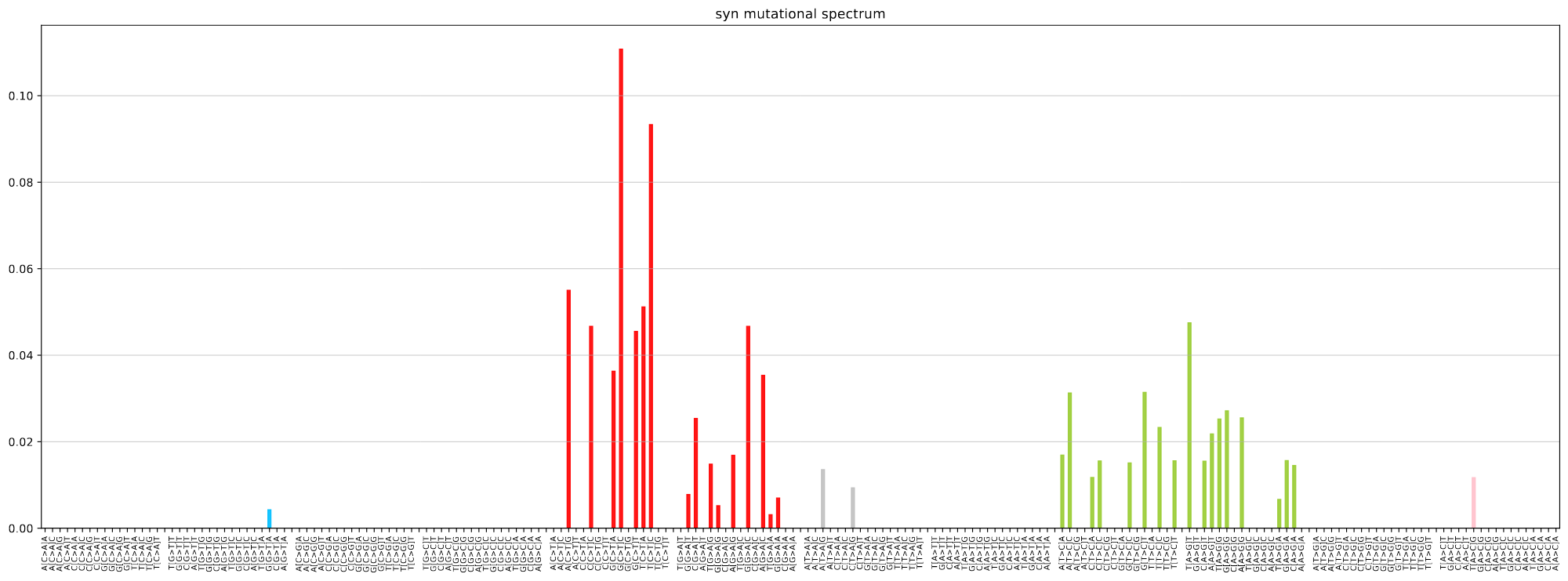
plot_mutspec192(spectra192)- IQ-Tree2 - efficient software for phylogenomic inference
- Genetic codes
Bogdan Efimenko, Konstantin Popadin, Konstantin Gunbin, NeMu: a comprehensive pipeline for accurate reconstruction of neutral mutation spectra from evolutionary data, Nucleic Acids Research, Volume 52, Issue W1, 5 July 2024, Pages W108–W115, https://doi.org/10.1093/nar/gkae438