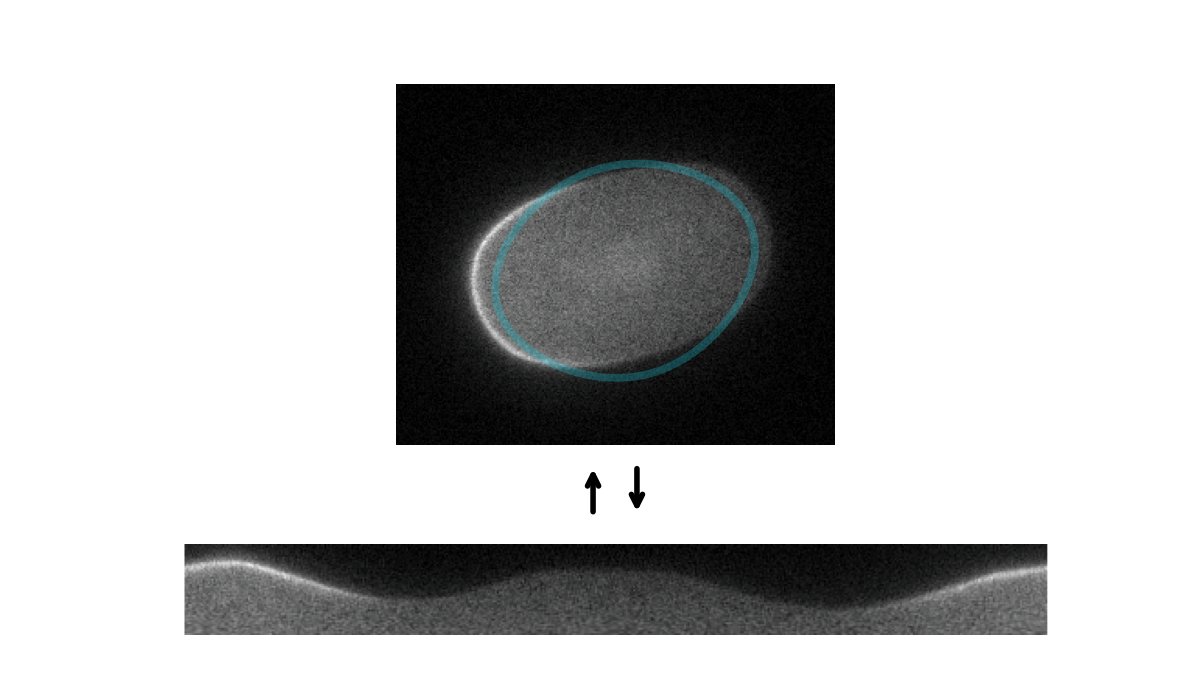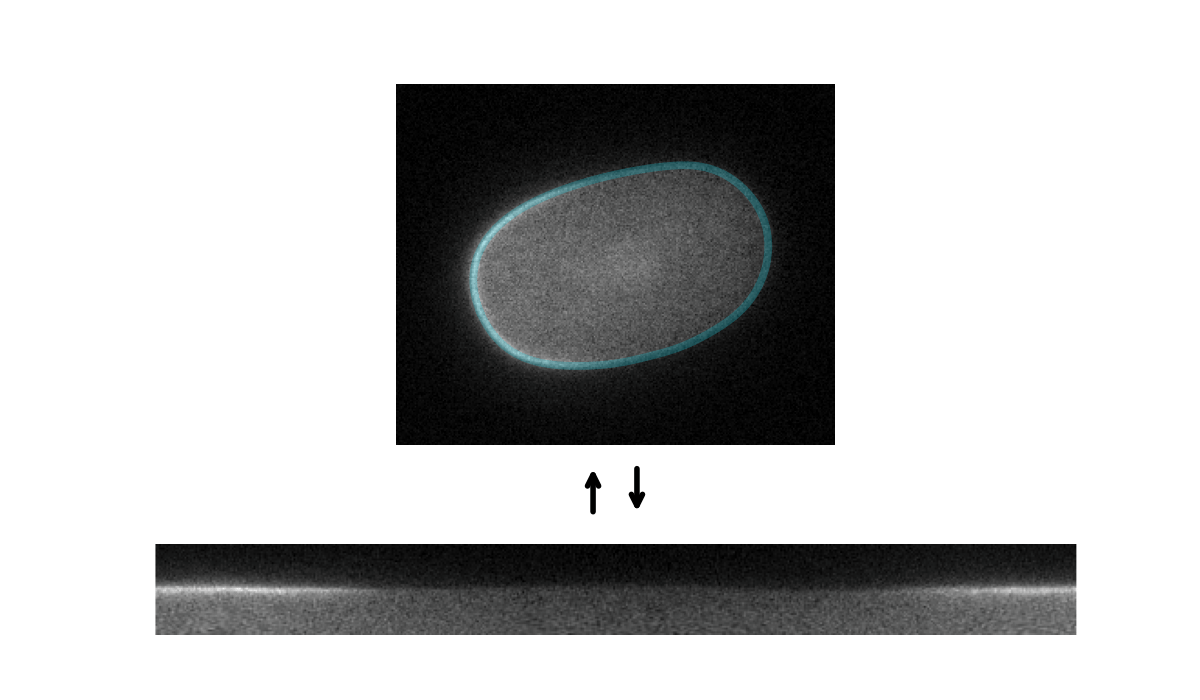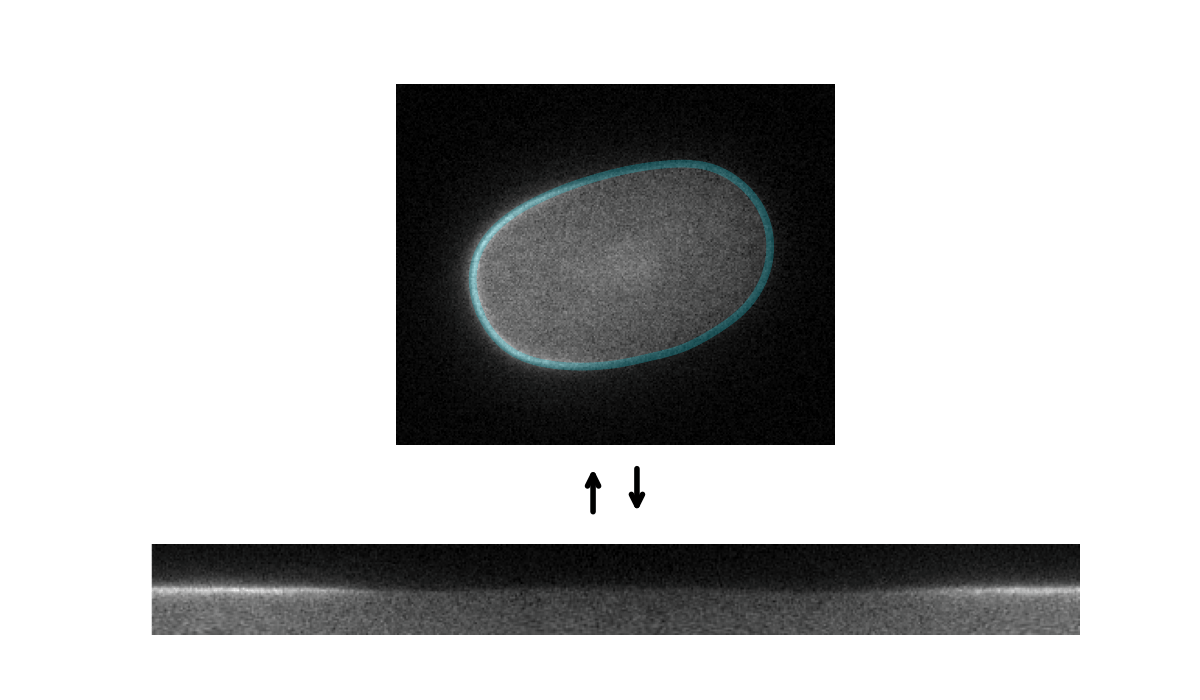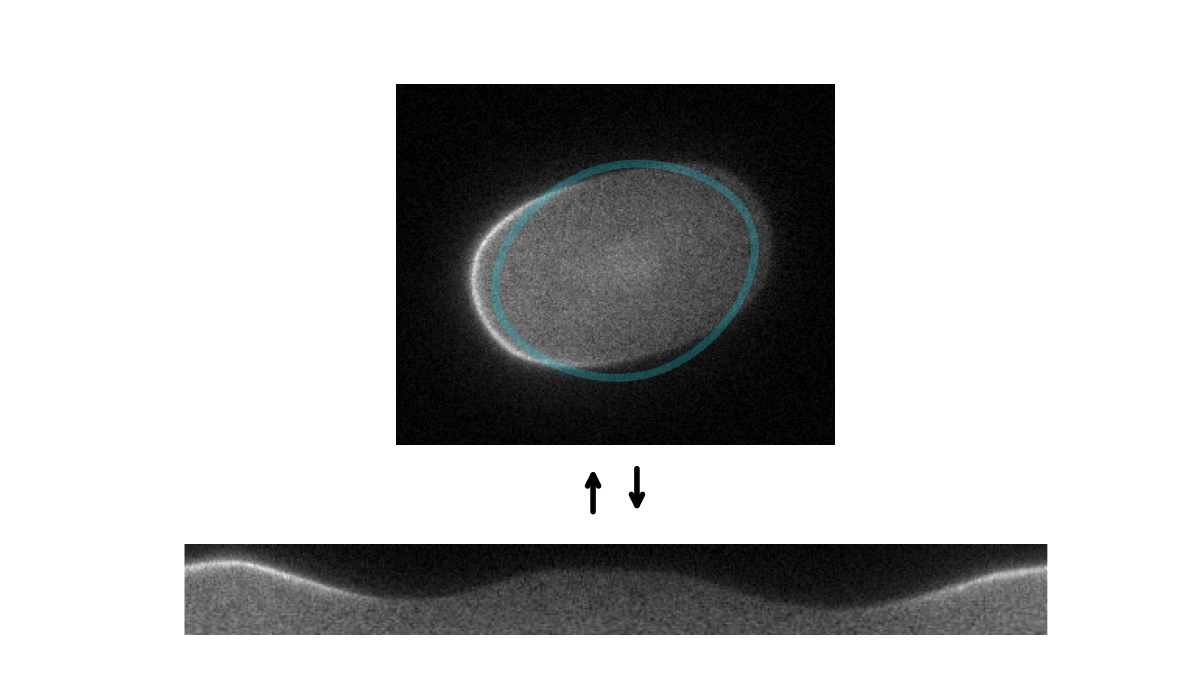
Tools for segmenting, straightening and quantifying the cortex of cells. Works by combining spline-based segmentation with a custom quantification model, using a gradient descent optimisation procedure. Designed primarily for membrane-bound PAR proteins in C. elegans zygotes.
Advantages:
- Combine segmentation and membrane quantification in a single step
- No ground truth training data required
- Works for single snapshots or timelapse movies
Disadvantages:
- Requires a small amount of manual annotation for every image
- A little slow compared to some other segmentation methods
As a first step, I would recommend checking out the tutorial notebook. This can be run in the cloud using Binder (please note that it may take several minutes to open the notebook):
To run locally, download the code and install the relevant requirements (requirements.txt) in a virtual environment.
To explore further and incorporate into your own analysis pipelines, you can install the package from PyPI using pip:
pip install par-segmentation
Depending on the underlying model you chose to use, you may have to install additional dependencies:
pip install par-segmentation[tensorflow,jax]
If you want to make changes to the code, you can download/clone this repository, navigate to it, and run:
pip install -e .[all]
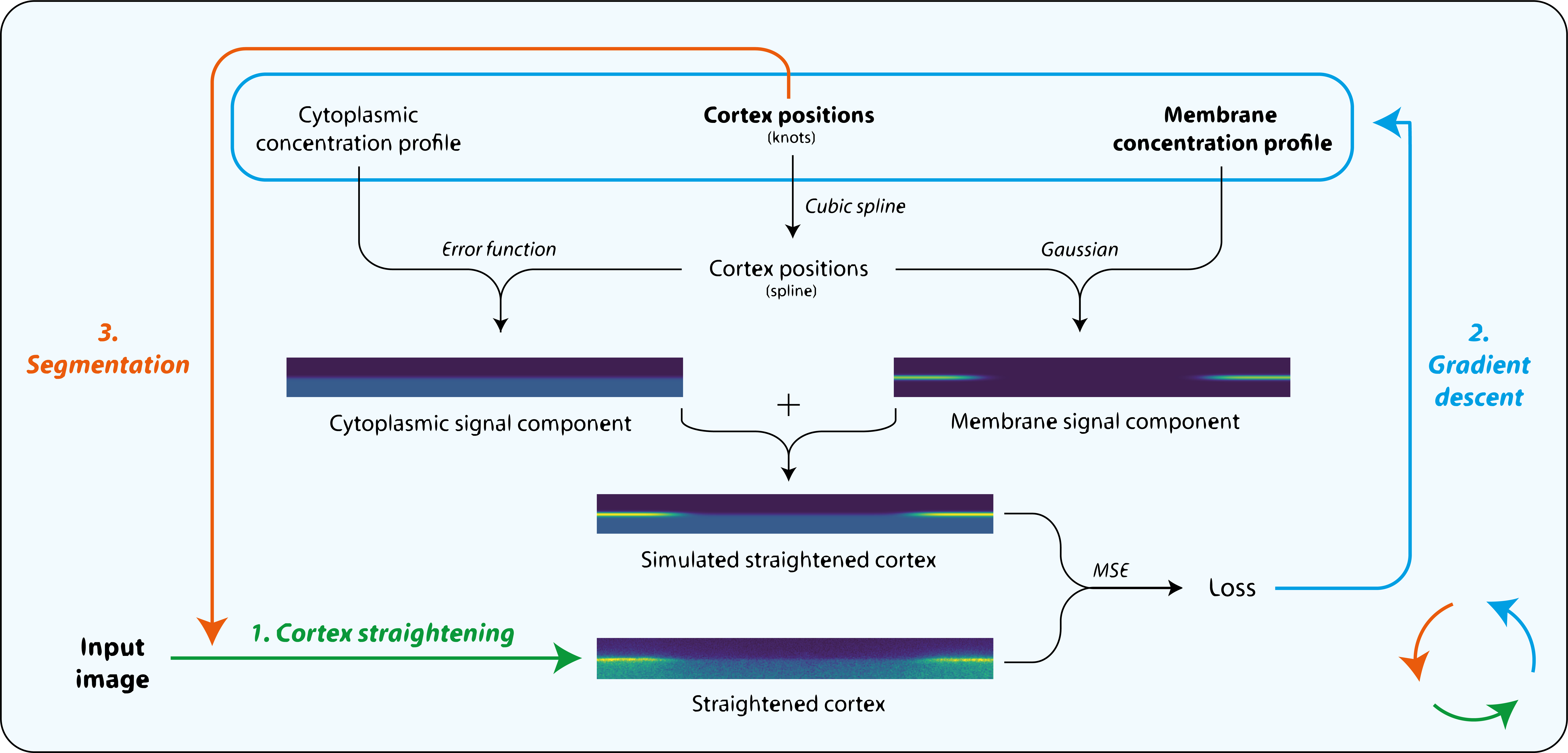
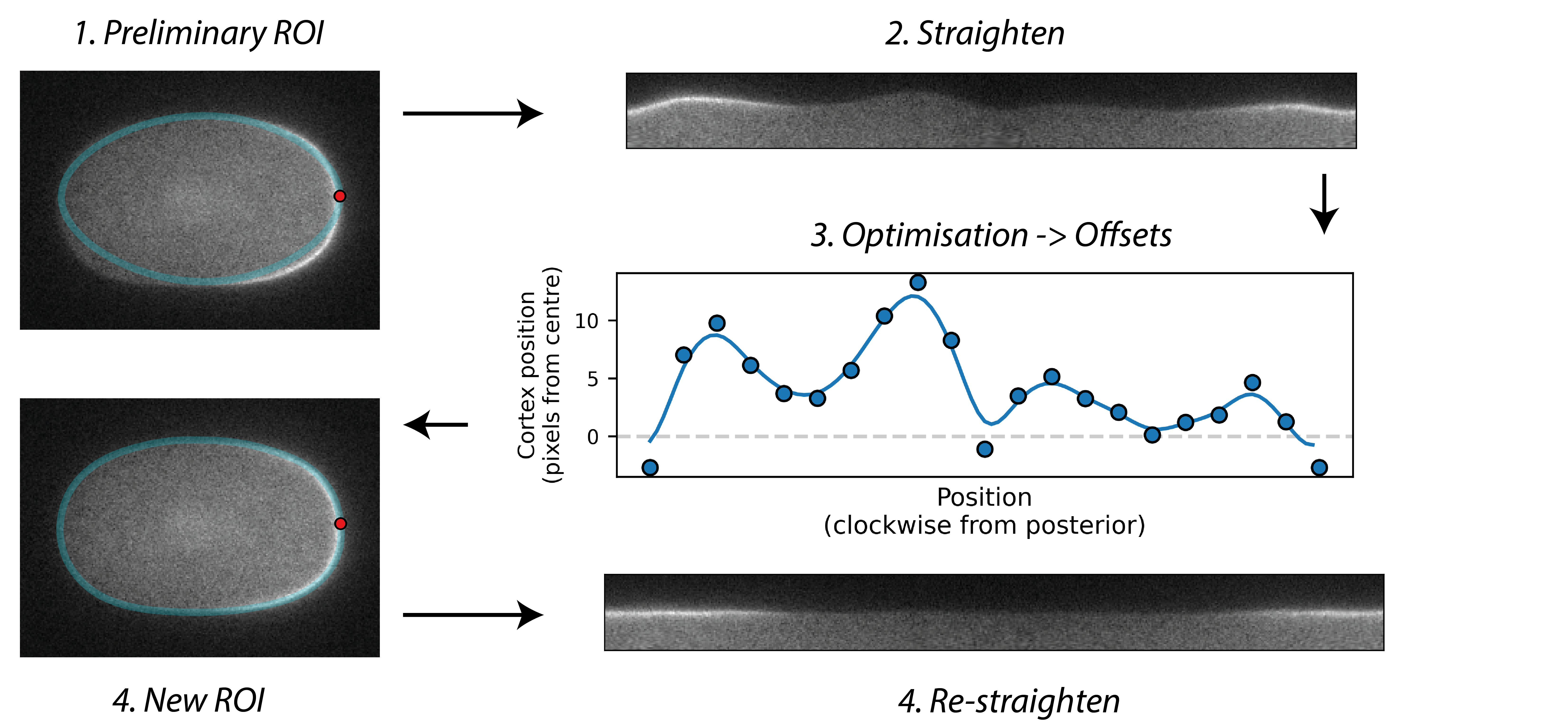
Starting with an initial rough manual ROI of the cell edge, the cortex of the image is straightened (Step 1). The program then attempts to mimic this straightened image by differentiable simulation (Step 2). In doing so, it learns the position of the cortex, which enables the ROI to be adjusted (Step 3) and the cortex re-straightened.
Cortex positions are modelled as a spline with a user-specified number of evenly spaced knots which are optimised by gradient descent:
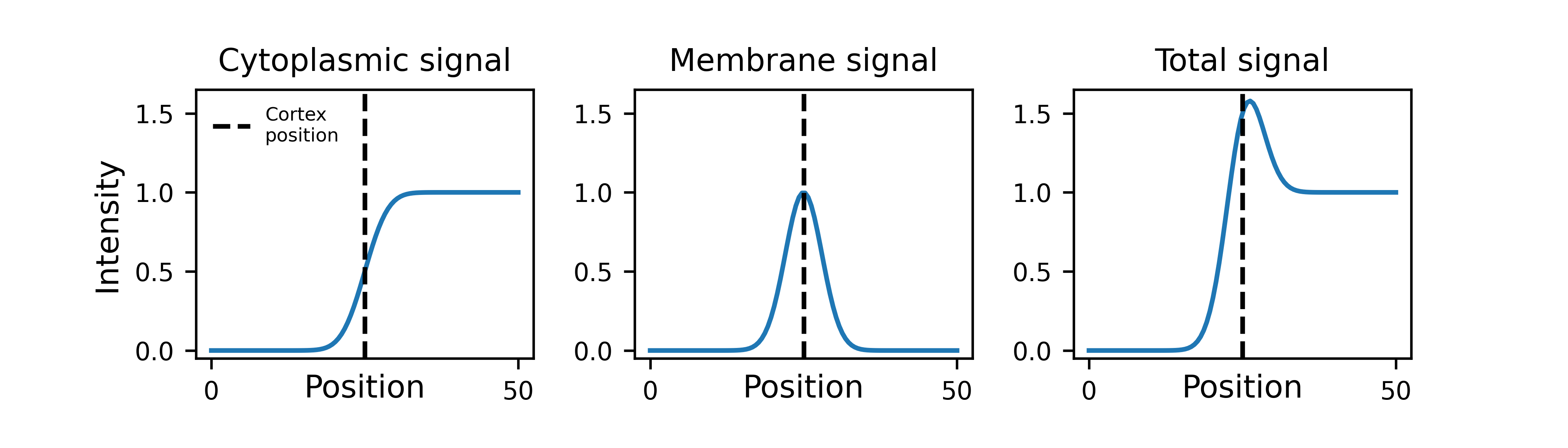
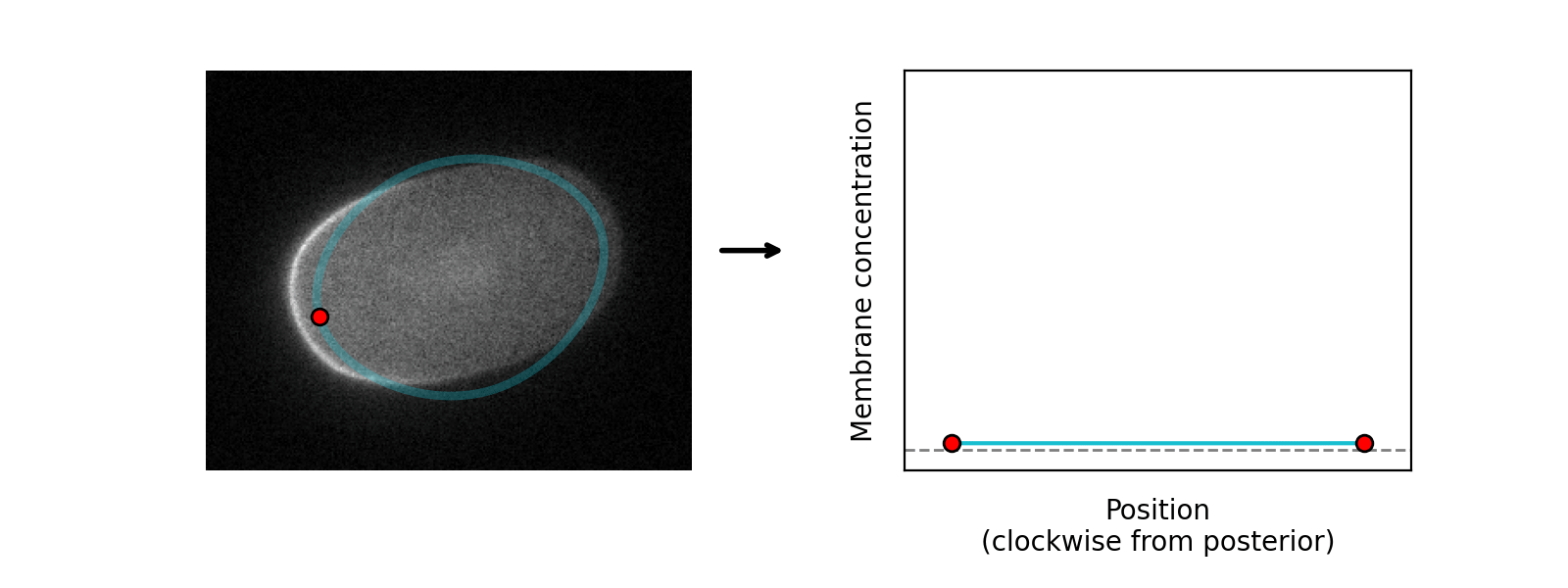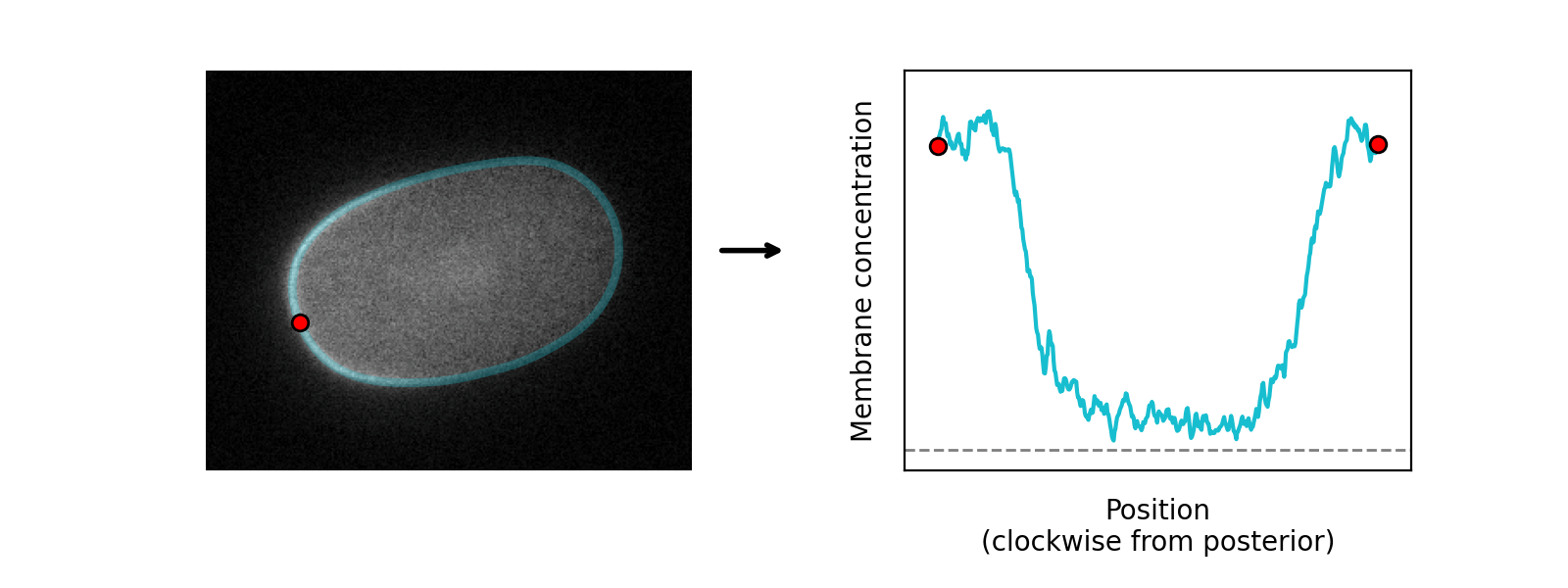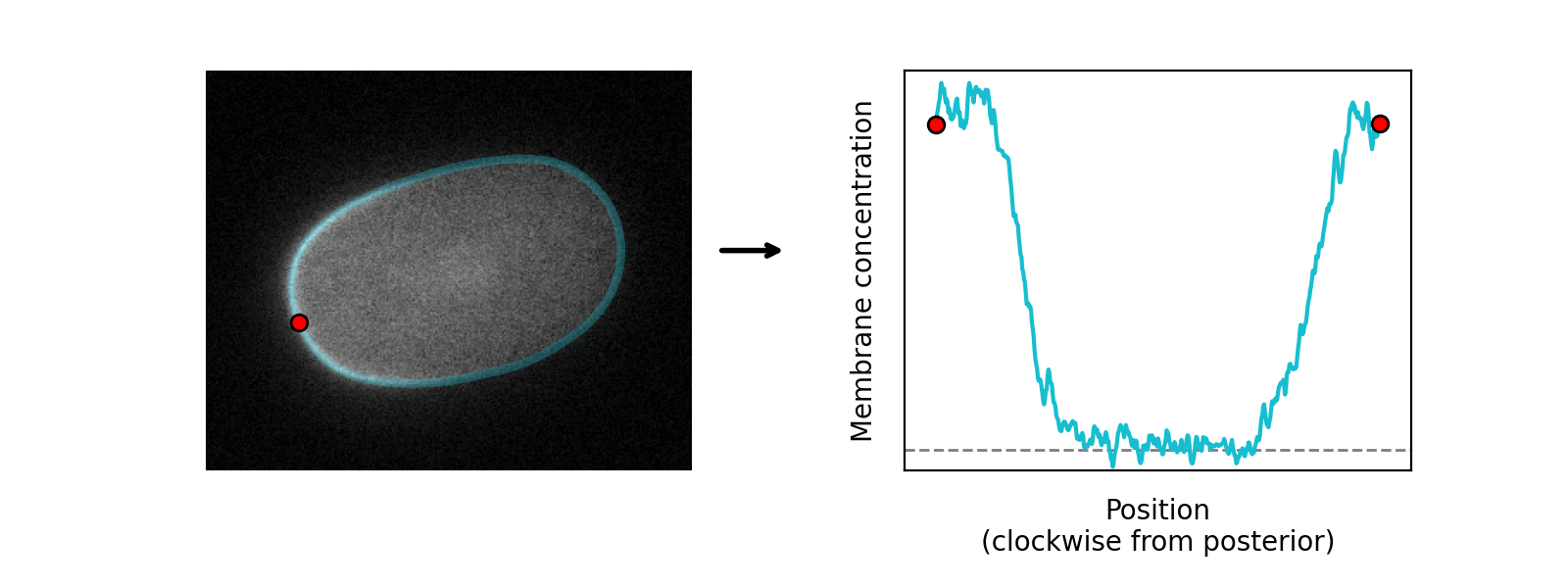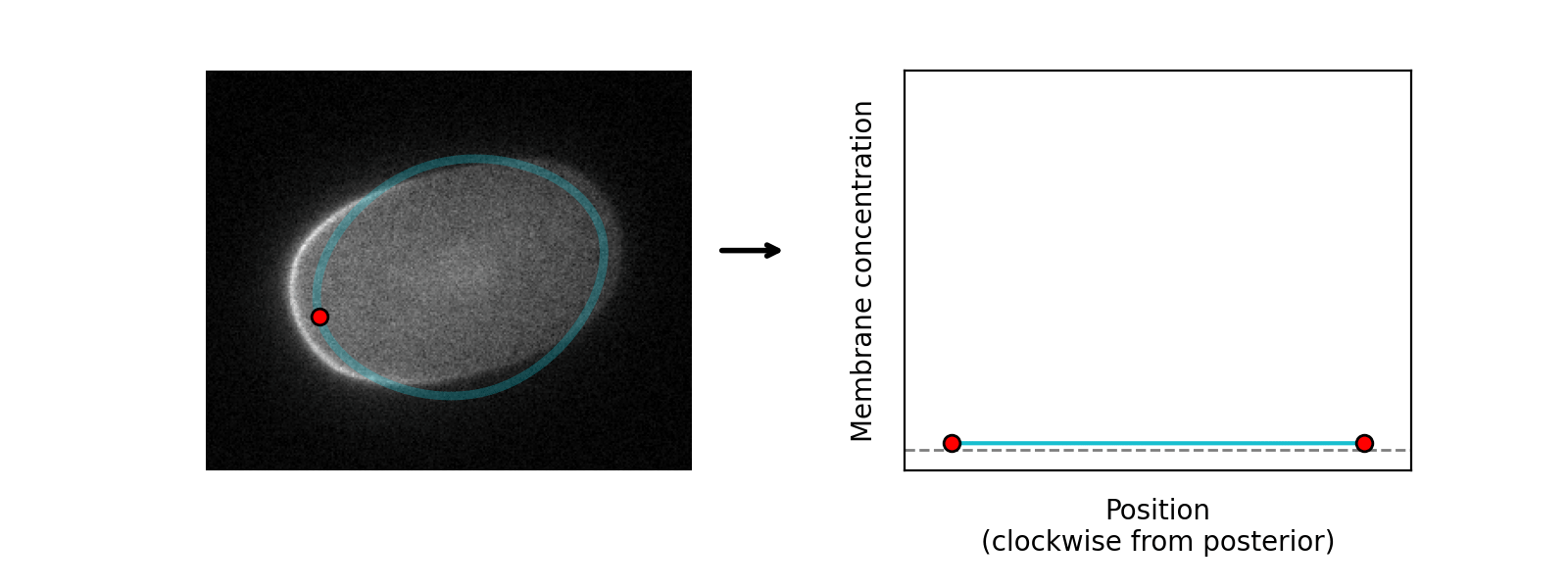
In the default model, cross-cortex intensity profiles at each position around the cortex are modelled as the sum of distinct cytoplasmic and membrane signal components: an error function and Gaussian function respectively, representing the expected shape of a step and a point convolved by a Gaussian point spread function (PSF) in one dimension:
The program learns the amplitude of these two components at each position around the cortex, so can serve as a quantification tool as well as a segmentation tool:
An additional model is included that can relax these assumptions if higher accuracy is required, see here
This package has been used in the following publications:
-
Design principles for selective polarization of PAR proteins by cortical flows. Journal of Cell Biology (2023).
-
Cleavage furrow-directed cortical flows bias mechanochemical pathways for PAR polarization in the C . elegans germ lineage. BioRxiv (2022).
-
An analog sensitive allele permits rapid and reversible chemical inhibition of PKC-3 activity in C. elegans. Micropublication Biology (2022).
-
SAIBR : A simple, platform-independent method for spectral autofluorescence correction. Development (2022).
To add your paper to this list, please use the issues form, or create a pull request
This work is licensed under a Creative Commons Attribution 4.0 International License.